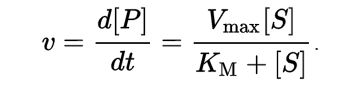Part:BBa_M1436:Experience
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_M1436
User Reviews
Group: Virginia iGEM 2016
Authors: Daniel Katz, Raquel Moya
Summary: In order to determine the relative efficiency of our enzyme (BBa_K1879001), we modeled another BioBrick. BBa_M1436 codes for pepsinogen, an inactive form of the cleavage enzyme pepsin. Pepsin is a nonspecific cleavage enzyme that cleaves dipeptide bonds between amino acids. Comparisons between pepsin and our enzyme may yield interesting results, specifically regarding the relationship of substrate concentration as it varies over time.
Documentation:
Results
Modeling was conducted by solving for Michaelis-Menten equations in MatLab. Both Equilibrium (EA) and Quasi-Steady-State (QSSA) Approximation were used to modeling reaction rates. To determine which approach would have the most accurate results, we analysed the two error equations:
for EA
for QSSA
Once analysed, it became aparent that the error term of EA would always be greater than one, while the error term of QSSA was sometimes within bounds. Future graphs do not display values that fall outside of the established error ranges.
Varying concentrations of substrate and enzyme were run through the QSSA equation:
Results were visualized on two concentration ranges, 0 to 10 mM and 0 to 1 mM. This was influenced mainly by the fact that, as [E] decreased, more values fell within error bounds.
Please see our modeling page for the full results and figures. http://2016.igem.org/Team:Virginia/Model
UNIQea942ab50b18e576-partinfo-00000000-QINU
UNIQea942ab50b18e576-partinfo-00000001-QINU